-
-
Notifications
You must be signed in to change notification settings - Fork 18.4k
BUG: to_datetime very slow with unsigned ints for unix seconds #42606
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Comments
|
this is fixed for 1.3 |
|
I still see this with pandas pandas 1.3.0 and numpy 1.20.3. |
|
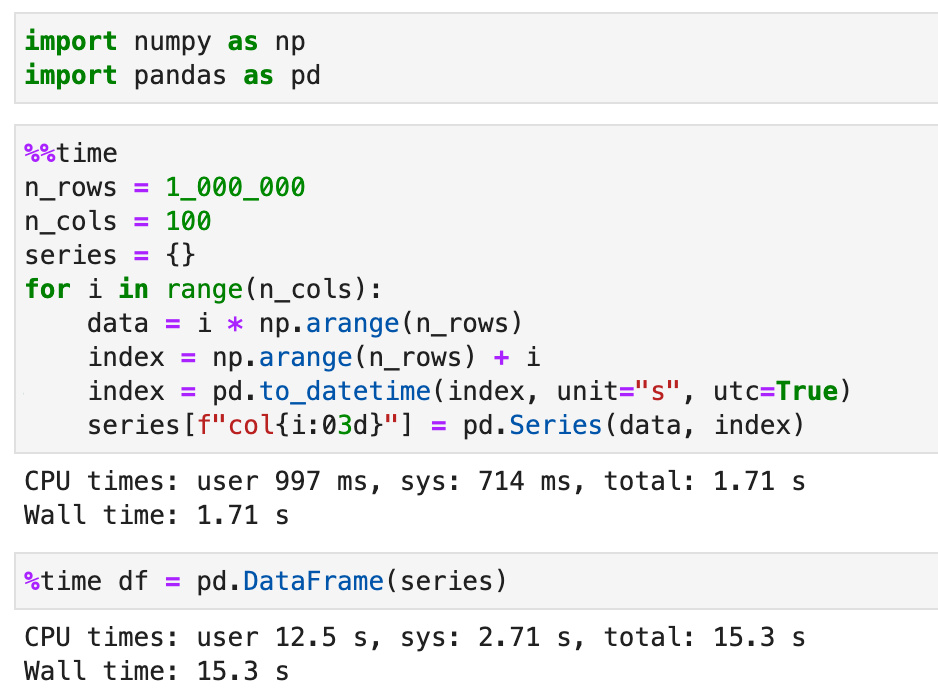
After changing my pipeline to use signed integers, the bottleneck then becomes joining ~ 100 Series with a few million data points and sorted datetime index, as in this example: n_rows = 1_000_000
n_cols = 100
series = {}
for i in range(n_cols):
data = i * np.arange(n_rows)
index = np.arange(n_rows) + i
index = pd.to_datetime(index, unit="s", utc=True)
series[f"col{i:03d}"] = pd.Series(data, index)
df = pd.DataFrame(series)Is calling In my real code with IoT sensor time series the indexes are int unix seconds and sorted, but a little bit different for each of the time series, so I need some kind of join / concat. |
|
Profiling shows: that the whole time is spent in |
|
Here is line_profiling of Note: However, the issue compared to fast Lines 246 to 251 in ef99443
because for |
I have checked that this issue has not already been reported.
I have confirmed this bug exists on the latest version of pandas.
(optional) I have confirmed this bug exists on the master branch of pandas.
I had a data pipeline that was terribly slow. Turns out the issue was that all the time was spent in pd.to_datetime calls with unix sec integers as input, because I was passing big-endian ints. With normal ints it's about 1000x faster.
Can anyone reproduce this performance issue?
is it possible to improve on this "gotcha", e.g. by forcing a typecast on input, or even some other way that doesn't require a copy and extra memory?
Output of
pd.show_versions()In [11]: pd.show_versions()
INSTALLED VERSIONS
commit : 2cb9652
python : 3.8.10.final.0
python-bits : 64
OS : Darwin
OS-release : 20.5.0
Version : Darwin Kernel Version 20.5.0: Sat May 8 05:10:33 PDT 2021; root:xnu-7195.121.3~9/RELEASE_X86_64
machine : x86_64
processor : i386
byteorder : little
LC_ALL : None
LANG : None
LOCALE : None.UTF-8
pandas : 1.2.4
numpy : 1.20.3
pytz : 2021.1
dateutil : 2.8.1
pip : 21.1.2
setuptools : 49.6.0.post20210108
Cython : None
pytest : 6.2.4
hypothesis : None
sphinx : None
blosc : None
feather : None
xlsxwriter : None
lxml.etree : None
html5lib : None
pymysql : None
psycopg2 : 2.8.6 (dt dec pq3 ext lo64)
jinja2 : 3.0.1
IPython : 7.24.1
pandas_datareader: None
bs4 : 4.9.3
bottleneck : 1.3.2
fsspec : 2021.05.0
fastparquet : None
gcsfs : None
matplotlib : 3.4.2
numexpr : None
odfpy : None
openpyxl : 3.0.7
pandas_gbq : None
pyarrow : 4.0.1
pyxlsb : None
s3fs : None
scipy : 1.6.3
sqlalchemy : 1.4.18
tables : None
tabulate : 0.8.9
xarray : 0.18.2
xlrd : 1.2.0
xlwt : None
numba : 0.53.1
The text was updated successfully, but these errors were encountered: